In the world of marine biology and ecology, the diversity of zooplankton presents a unique challenge. These tiny organisms vary in species, life stages, and ecological roles. To tackle this complexity, we’ve adopted a polyhierarchical taxonomy approach, offering a versatile and nuanced system for zooplankton categorization.
Zooplankton often presents researchers with images that defy simple categorization. These images may reveal various aspects of life, including diverse species, life stages, and ecological roles. Therefore, a taxonomy that can adequately capture this complexity is essential.
Motivation
The conventional flat class system did not suit our needs, due to the following limitations:
- A “Copepod with eggs” is also a “Copepod”. However, both are distinct concepts in the conventional system.
- Creating new classes for every combination of phylogenetic classification and further properties (e.g. viewing angle, sex, life stage, image defects, presence of ectoparasites, …) proved to be both cumbersome and impractical.
- The overlapping classes posed significant hurdles for machine learning. (Why is this “Copepod with eggs” not also a “Copepod”? Why is this “ciiistage” different from that “ciiistage”?)
The Polyhierarchical Taxonomy Approach
Our system combines a primary phylogenetic classification, such as family, genus, or species, with further positive and negative descriptors. These descriptors could include attributes, life stages, and behaviors.
The primary classification anchors each zooplankton object in the taxonomic tree. Positive descriptors assign certain characteristics to an object, while negative descriptors negate specific attributes. Negative descriptors allow us to specify what an object is not. They help in eliminating ambiguities by indicating characteristics that are absent. By combining phylogenetic classification with descriptors, we gain a comprehensive understanding of each object, including orientation, life stage, gut content and other life history traits.
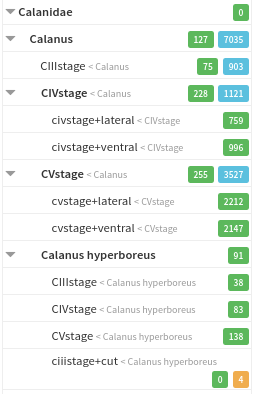
PolyTaxo introduces a polyhierarchical taxonomy that allows for a hierarchy of classes. For each class, tags can be defined. These tags are valid for all descendants of this class, e.g. a tag view:lateral defined for the class Copepoda is also valid for Copepoda/Calanus.
Example Taxonomy
# The root node of the class hierarchy
::
# These tags can be applied to all kinds of objects
cut:
multiple:
Copepoda::
# These tags can be applied to all Copepoda and subclasses
sex:
male:
female:
view:
lateral:
dorsal:
ventral:
ovigerous:
stage:
CI:
CII:
CIII:
CIV:
CV:
ectoparasites:
epibiont:
# Subclasses of Copepoda
Calanus::
Calanus finmarchicus::
Calanus hyperboreus::
Calanus glacialis::
Calanus other::
Metridia::
Metridia longa::
Metridia other::
Other Copepoda::
Other::
Object descriptions
Objects can be described at varying levels of specificity. A description is a string consisting of a primary classification and further positive and negative descriptors. If a tag is not specified in the description, its state (positive or negative) is undefined.
Copepoda sex:female: A female copepod.Calanus view:lateral epibiont: A Calanus seen from the side with an epibiont.Calanus !'Calanus hyperboreus': A Calanus of unknown species, but in all cases not a C. hyperboreus.
Object descriptions also form subset-relationshipts. For instance, Calanus is a subset of Copepoda, and Calanus view:lateral is a subset of Copepoda view:lateral. This allows to effectively query a dataset. For example, we need to find all descriptions that are a subset of Copepoda view:lateral and not a subset of cut.
Use in Machine Learning
The richness of description offered by PolyTaxo allows us to train multi-label classifiers with outputs for each primary (class) and secondary concept (tag). We expect this to not only improve accuracy but also to allow for the use of data for training that lacks certain annotations.
The MAZE-IPP Project
PolyTaxo was developed within the context of the „MOSAiC Zooplankton Image Analysis“ (MAZE) project, a collaborative effort between the Alfred-Wegener-Institut, the Christian-Albrechts-Universität zu Kiel, and the Laboratoire d’Océanographie de Villefranche-sur-Mer. It is dedicated to developing an image processing pipeline (MAZE-IPP). MAZE-IPP is designed to automate the assignment of taxonomic categories, such as orders, genera, species, and even developmental stages, to organisms in images. This will enable the calculation of crucial ecological parameters, including abundance, biomass, and respiration rates, from the resulting data.
The Road Ahead
PolyTaxo is an approach still in development, and its full potential is yet to be realized. As it evolves, we anticipate numerous benefits, including improved data management, more precise ecological assessments, and enhanced machine learning applications.